“Conductive polymers, including polypyrrole (PPy), have been extensively explored to fabricate electrically conductive biomaterials for bioelectrodes and tissue engineering scaffolds. For their in vivo uses, a sterilization method without severe impairment of original material properties and performance is necessary. Gamma-ray radiation has been commonly applied for sterilization of medical products because of its simple and uniform sterilization without heat generation.[…]”*
In the article “Effective gamma-ray sterilization and characterization of conductive polypyrrole biomaterials” by Semin Kim et. al cited here, the authors describe the first study on gamma-ray sterilization of PPy bioelectrodes and its effects on their characteristics.
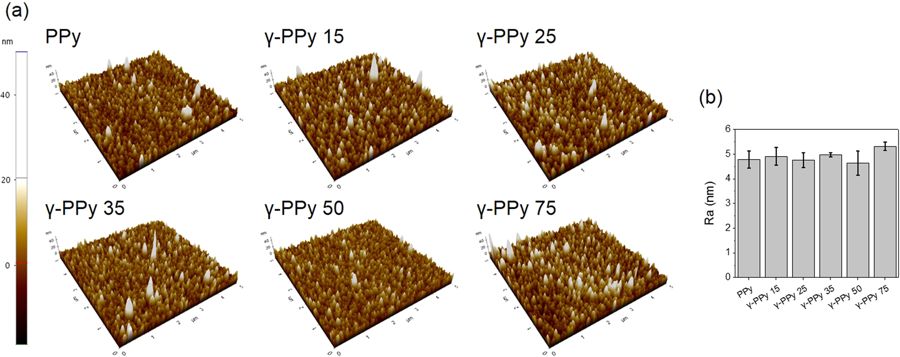
The surface topography and roughness of the PPy and γ-PPy electrodes were analyzed by atomic force microscopy. The experiments were performed using a NanoWorld Pointprobe® NCHR AFM probe. All images were acquired at a 0.3 Hz scan rate in tapping mode.
*Semin Kim, Jin-Oh Jeong, Sanghun Lee, Jong-Seok Park, Hui-Jeong Gwon, Sung In Jeong, John George Hardy, Youn-Mook Lim, Jae Young Lee
Effective gamma-ray sterilization and characterization of conductive polypyrrole biomaterials
Nature Scientific Reports, volume 8, Article number: 3721 (2018)
DOI: https://doi.org/10.1038/s41598-018-22066-6
Please follow this external link for the full article: https://rdcu.be/bariF
Open Access: The article “Effective gamma-ray sterilization and characterization of conductive polypyrrole biomaterials” by Semin Kim et. al is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/.




