The products of the polyurethane (PU) industry such as foams, coatings and adhesives are numerous and can be found in many areas of everyday life. *
Polyols are an essential component in the production of polyurethane. Nowadays they mostly come from petroleum products. *
In view of potential risk factors such as the running out of fossil fuels, supply chain issues, environmental concerns and economic risks it is important to develop alternatives as substitutes and supplements to the existing petroleum derived polyols. *
Vegetable oils can be used to manufacture biobased polyols and various oils such as linseed oil, rapeseed oil, canola oil, grapeseed oil, corn oil, rice bran oil, palm oil, olive oil, castor oil and soybean oil have already been used to make polyols for different purposes.*
Most of the polyols derived from vegetable oils that are already commercially available are made from soybean and castor oil and are mainly used for rigid PU foam applications. *
So far biobased materials for flexible polyurethane foams (FPUFs) have not been studied as much as their rigid counterpart. This is because, due to their chemical composition, there are limits to how much biobased materials can be used in the flexible foam without having an undesired effect on the foam’s mechanical properties. *
Coconut oil is not often used to manufacture flexible foam because the coconut oil’s high level of saturation makes it less compatible with many common methods of creating polyols, as the widely used polyol-forming processes mostly rely on the unsaturation of vegetable oil for functionalization.
To make coconut monoglycerides (CMG) or other plant-based oils usable for polyol-forming processes they need to fulfil the same structural requirements as the fossil-based products. *
In the article “Flexible Polyurethane Foams Modified with Novel Coconut Monoglycerides-Based Polyester Polyols “ Christine Joy M. Omisol, Blessy Joy M. Aguinid, Gerson Y. Abilay, Dan Michael Asequia, Tomas Ralph Tomon, Karyl Xyrra Sabulbero, Daisy Jane Erjeno, Carlo Kurt Osorio, Shashwa Usop, Roberto Malaluan, Gerard Dumancas, Eleazer P. Resurreccion, Alona Lubguban, Glenn Apostol, Henry Siy, Arnold C. Alguno, and Arnold Lubguban describe how they investigated the potential of coconut monoglycerides (CMG) as a polyol raw material specifically for flexible polyurethane foam (FPUF) applications.*
The authors synthesized high-molecular-weight polyester polyols from coconut monoglycerides (CMG), a coproduct of fatty acid production from coconut oil, via polycondensation at different mass ratios of CMG with 1:5 glycerol:phthalic anhydride.*
The resulting CMG-based polyols were shown to work well in making flexible foam. *
Fourier transform infrared (FTIR) spectroscopy and atomic force microscopy (AFM) were used for the foam characterization. *
The modification of the foam formulation increased the monodentate and bidentate urea groups, shown using Fourier transform infrared (FTIR) spectroscopy, that promoted microphase separation in the foam matrix, confirmed using atomic force microscopy (AFM) and differential scanning calorimetry (DSC). *
Atomic force microscopy (AFM) was used to evaluate the hard–soft domains phase separation of the foam. *
The atomic force microscope was operated at a scan rate of 1.0 Hz in non-contact mode using NanoWorld Pointprobe® NCHR Silicon AFM probes for standard tapping mode applications. (typical resonance frequency 320 kHz, typical force constant 42 N/m). *
It could be shown that density of the CMGPOL-modified polyurethane foams (CPFs) decreased, while a significant improvement in their tensile and compressive properties was observed. *
The investigations by Christine Joy M. Omisol et al. resulted in a new sustainable polyol raw material that can be used to modify petroleum-based foam and produce flexible foams with varying properties that can be tailored to meet specific requirements. *
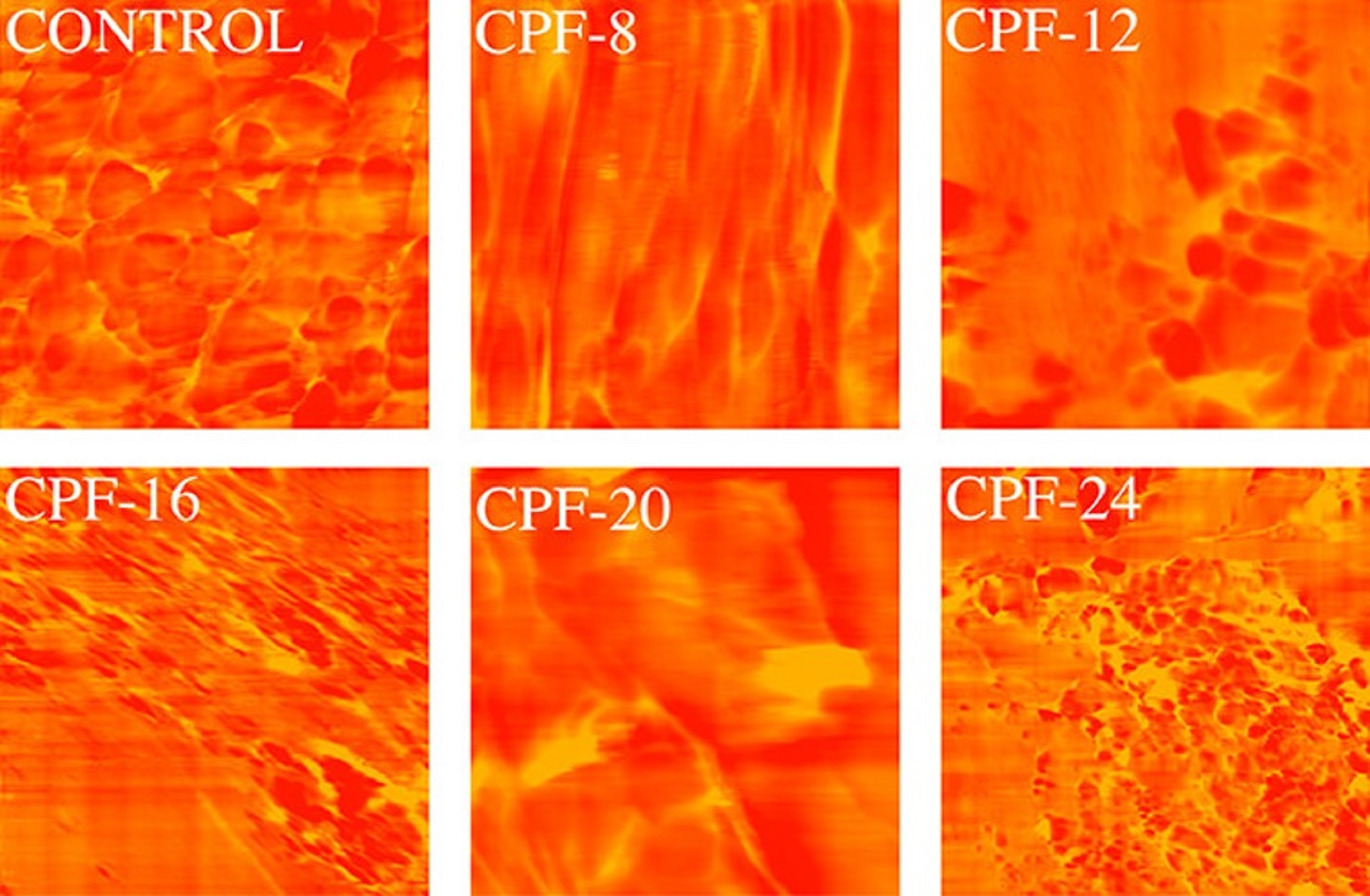
Atomic force microscopy (AFM) phase images of CMGPOL-modified polyurethane foams (CPF) and control foam measured with a size scan of 3 μm × 3 μm showing soft and hard regions represented by red and yellow colors, respectively.
*Christine Joy M. Omisol, Blessy Joy M. Aguinid, Gerson Y. Abilay, Dan Michael Asequia, Tomas Ralph Tomon, Karyl Xyrra Sabulbero, Daisy Jane Erjeno, Carlo Kurt Osorio, Shashwa Usop, Roberto Malaluan, Gerard Dumancas, Eleazer P. Resurreccion, Alona Lubguban, Glenn Apostol, Henry Siy, Arnold C. Alguno, and Arnold Lubguban
Flexible Polyurethane Foams Modified with Novel Coconut Monoglycerides-Based Polyester Polyols
ACS Omega 2024, 9, 4, 4497–4512
DOI: https://doi.org/10.1021/acsomega.3c07312
The article “Flexible Polyurethane Foams Modified with Novel Coconut Monoglycerides-Based Polyester Polyols” by Christine Joy M. Omisol, Blessy Joy M. Aguinid, Gerson Y. Abilay, Dan Michael Asequia, Tomas Ralph Tomon, Karyl Xyrra Sabulbero, Daisy Jane Erjeno, Carlo Kurt Osorio, Shashwa Usop, Roberto Malaluan, Gerard Dumancas, Eleazer P. Resurreccion, Alona Lubguban, Glenn Apostol, Henry Siy, Arnold C. Alguno and Arnold Lubguban is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third-party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/.


![Figure 15 from Ilsiya M. Davletbaeva et al “Amphiphilic Poly(dimethylsiloxane-ethylene-propylene oxide)-polyisocyanurate Cross-Linked Block Copolymers in a Membrane Gas Separation”:
AFM Images. (a): [PPEG]:[TDI] = 1:10; (b): [PPEG]:[D4]:[TDI] = 1:15:10; (c): [PPEG]:[D4]:[TDI] = 1:15:10 [ASiP] = 0.2 wt.%, (d): [PPEG]:[D4]:[TDI] = 1:15:10 [ASiP] = 0.4 wt.%.
NanoWorld Pointprobe® FMR AFM probes were used.](https://dhipgo7nn2tea.cloudfront.net/wp-content/uploads/2021/02/08182612/figure-15-from-I-Davletbaeva-et-al-Amphiphilic-Polydimethylsiloxane-ethylene-propylene-oxide-polyisocyanurate-Cross-Linked-Block-Copolymers-in-a-Membrane-Gas-Separation-Pointprobe-FMR-AFM-probe.jpg)