The emergence of multiple antibiotic-resistant bacteria, notably, pan-resistant Gram-negative pathogens, which are equipped with an outer membrane barrier of low permeability to antibiotics, has become an important challenge in recent decades following the overuse of antibiotics in humans and animals. *
In particular, the foodborne enteric pathogen Escherichia coli O157:H7 has caused severe or deadly illness cases worldwide. *
Among E. coli O157 isolates, serotype O157:H7 is the most common enteric pathogen isolated from patients with bloody diarrhea and it is also frequently found in non-bloody diarrhea samples. Many of its clinical isolates from humans and animals as well as isolates from contaminated food have been found to develop resistance to several antibiotics. *
Following the first isolation of mastoparan, the most abundant peptide in the hornet or wasp venom, from Vespula lewisii, many homologs of mastoparan were isolated from various hornets and solitary wasps. *
Mastoparan homologs are cationic tetradecapeptides with membrane permeabilizing activity and antimicrobial activity on various bacteria, mast cell degranulation activity, and hemolytic activity. *
In the article “Antimicrobial Peptide Mastoparan-AF Kills Multi-Antibiotic Resistant Escherichia coli O157:H7 via Multiple Membrane Disruption Patterns and Likely by Adopting 3–11 Amphipathic Helices to Favor Membrane Interaction” Chun-Hsien Lin, Ching-Lin Shyu, Zong-Yen Wu, Chao-Min Wang, Shiow-Her Chiou, Jiann-Yeu Chen, Shu-Ying Tseng, Ting-Er Lin, Yi-Po Yuan, Shu-Peng Ho, Kwong-Chung Tung, Frank Chiahung Mao, Han-Jung Lee and Wu-chun Tu investigate the antimicrobial activity and membrane disruption modes of the antimicrobial peptide mastoparan-AF against hemolytic Escherichia coli O157:H7.*
Based on the physicochemical properties, mastoparan-AF may potentially adopt a 3–11 amphipathic helix-type structure, with five to seven nonpolar or hydrophobic amino acid residues forming the hydrophobic face. E. coli O157:H7 and two diarrheagenic E. coli veterinary clinical isolates, which are highly resistant to multiple antibiotics, are sensitive to mastoparan-AF, with minimum inhibitory and bactericidal concentrations (MIC and MBC) ranging from 16 to 32 μg mL−1 for E. coli O157:H7 and four to eight μg mL−1 for the latter two isolates. *
Mastoparan-AF treatment, which correlates proportionally with membrane permeabilization of the bacteria, may lead to abnormal dents, large perforations or full opening at apical ends (hollow tubes), vesicle budding, and membrane corrugation and invagination forming irregular pits or pores on E. coli O157:H7 surface. In addition, mRNAs of prepromastoparan-AF and prepromastoparan-B share a 5′-poly(A) leader sequence at the 5′-UTR known for the advantage in cap-independent translation. *
This is the first report about the physicochemical adaptation of 3–11 amphipathic helices among mastoparans or antimicrobial peptides. *
Considering that E. coli O157:H7 and clinical isolates are highly resistant to multiple classes of antibiotics, mastoparan-AF, with little or mild effect on animal RBCs, could be an effective and alternative treatment to combat hemolytic E. coli O157:H7 and other pathogenic E. coli.*
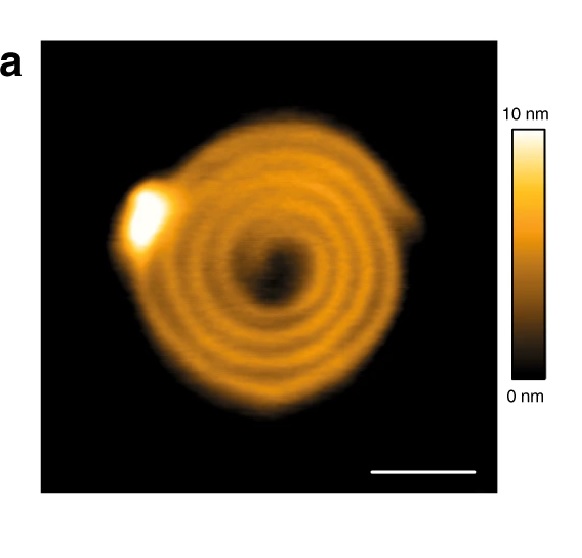
The topography of bacteria was measured by a commercial atomic force microscope using NanoWorld Pointprobe® NCSTR AFM probes with a typical resonance frequency of 160 kHz and a typical spring constant of 7.4 N/m, respectively. For image quality, the scan rates of the tip were 0.3–0.6 Hz, with a resolution set of 512 by 256 pixels, and the feedback control parameters were optimized. *

The topology of mastoparan-AF treated-hemolytic E. coli O157:H7 analyzed by AFM. (A) Two-dimensional (2D) and (B) three-dimensional (3D) images show smooth cell surfaces of untreated hemolytic E. coli O157:H7. (C) A 2D image of mastoparan-AF (32 μg mL−1)-treated hemolytic E. coli O157:H7. Abnormal perforations and dents on the surface of bacteria are indicated by arrows and arrowheads, respectively. The 3D images focusing on two highlighted areas of (C), respectively, reveal (D) a rough cell surface and (E) a hollow tube resulting from perforations at apical ends. (F) A 3D image shows a mastoparan-AF-treated bacterium with a budding vesicle. (G) A 3D image shows mastoparan-AF-treated bacteria with a wrinkled or rough surface. (H) Magnification of portion of (G) displays, in high resolution, the surface roughness of a mastoparan-AF-treated bacterium.
*Chun-Hsien Lin, Ching-Lin Shyu, Zong-Yen Wu, Chao-Min Wang, Shiow-Her Chiou, Jiann-Yeu Chen, Shu-Ying Tseng, Ting-Er Lin, Yi-Po Yuan, Shu-Peng Ho, Kwong-Chung Tung, Frank Chiahung Mao, Han-Jung Lee and Wu-chun Tu
Antimicrobial Peptide Mastoparan-AF Kills Multi-Antibiotic Resistant Escherichia coli O157:H7 via Multiple Membrane Disruption Patterns and Likely by Adopting 3–11 Amphipathic Helices to Favor Membrane Interaction
Membranes 2023, 13(2), 251
DOI: https://doi.org/10.3390/membranes13020251
The article “Antimicrobial Peptide Mastoparan-AF Kills Multi-Antibiotic Resistant Escherichia coli O157:H7 via Multiple Membrane Disruption Patterns and Likely by Adopting 3–11 Amphipathic Helices to Favor Membrane Interaction” by Chun-Hsien Lin, Ching-Lin Shyu, Zong-Yen Wu, Chao-Min Wang, Shiow-Her Chiou, Jiann-Yeu Chen, Shu-Ying Tseng, Ting-Er Lin, Yi-Po Yuan, Shu-Peng Ho, Kwong-Chung Tung, Frank Chiahung Mao, Han-Jung Lee and Wu-chun Tu is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third-party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/.