Ceramides are central intermediates of sphingolipid metabolism that also function as potent messengers in stress signaling and apoptosis. Progress in understanding how ceramides execute their biological roles is hampered by a lack of methods to manipulate their cellular levels and metabolic fate with appropriate spatiotemporal precision.*
In the article “Optical manipulation of sphingolipid biosynthesis using photoswitchable ceramides” Matthijs Kol, Ben Williams, Henry Toombs-Ruane, Henri G Franquelim, Sergei Korneev, Christian Schroeer, Petra Schwille, Dirk Trauner, Joost CM Holthuis and James A Frank report on clickable, azobenzene-containing ceramides, caCers, as photoswitchable metabolic substrates to exert optical control over sphingolipid production in cells.*
They combine atomic force microscopy on model bilayers with metabolic tracing studies in cells, and demonstrate that light-induced alterations in the lateral packing of caCers lead to marked differences in their metabolic conversion by sphingomyelin synthase and glucosylceramide synthase. These changes in metabolic rates are instant and reversible over several cycles of photoswitching. The findings described in the article disclose new opportunities to probe the causal roles of ceramides and their metabolic derivatives in a wide array of sphingolipid-dependent cellular processes with the spatiotemporal precision of light.*
The High-speed AFM in AC mode described in the article was done with NanoWorld Ultra-Short Cantilevers USC-F0.3-k0.3 with a typical stiffness of 0.3 N/m. The AFM cantilever oscillation was tuned to a frequency of 100–150 kHz and the amplitude kept below 10 nm. The scan rate was set to 25–150 Hz. Images were acquired at 256 × 256 pixel resolution. All measurements were performed at room temperature. The force applied on the sample was minimized by continuously adjusting the set point and gain during imaging. Height, error, deflection and phase-shift signals were recorded and images were line-fitted as required.*
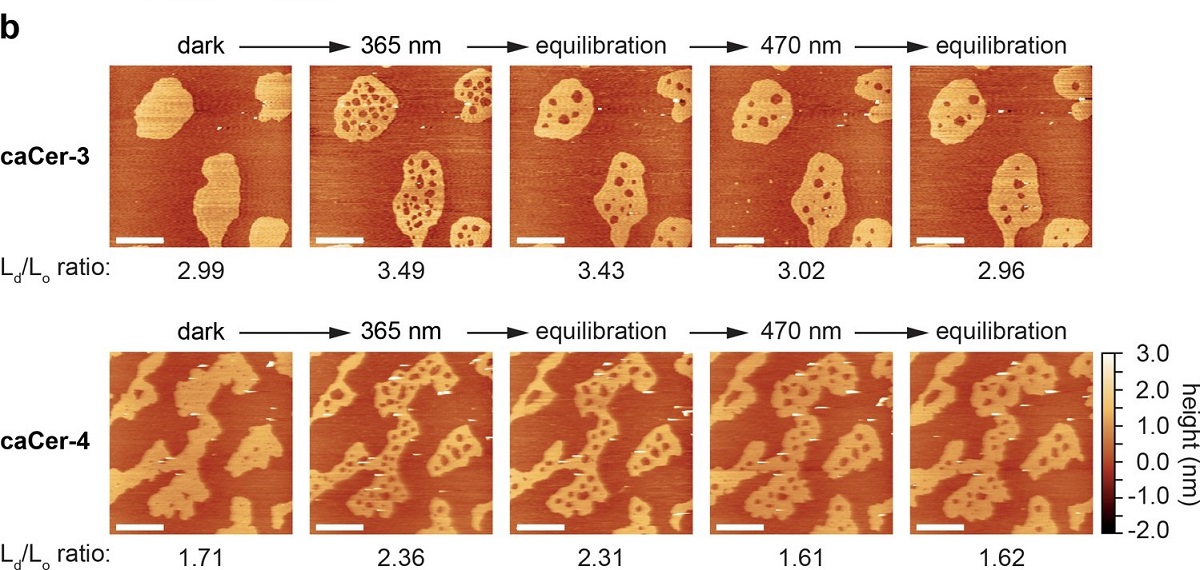
Photo-isomerization of caCers affects membrane fluidity and lipid domain structure in supported lipid bilayers.
(b) Atomic force microscopy of supported lipid bilayers ( SLBs )prepared as in (a). Isomerization of caCer-3 (top) and caCer-4 (bottom) to cis with UV-A light (365 nm) resulted in a fluidification inside the Lo domains, as indicated by the appearance of small fluid Ld lakes and an increased Ld/Lo area ratio. This effect was reversed on isomerization back to trans with blue light (470 nm), marked by a drop in the Ld/Lo area ratio. Scale bars, 2 μm. (c) Time-course plotting the normalized Lo area over multiple 365/470 nm irradiation cycles for caCer-3 (top) and caCer-4 (bottom).
Please refer to https://doi.org/10.7554/eLife.43230.007 for the full figure.
*Matthijs Kol, Ben Williams, Henry Toombs-Ruane, Henri G Franquelim, Sergei Korneev, Christian Schroeer, Petra Schwille, Dirk Trauner, Joost CM Holthuis, James A Frank
Optical manipulation of sphingolipid biosynthesis using photoswitchable ceramides
eLife 2019;8:e43230
DOI: 10.7554/eLife.43230
https://doi.org/10.7554/eLife.43230.001
https://doi.org/10.7554/eLife.43230.007
Please follow this external link to read the full article: https://elifesciences.org/articles/43230
Open Access The article “Optical manipulation of sphingolipid biosynthesis using photoswitchable ceramides “ by Matthijs Kol, Ben Williams, Henry Toombs-Ruane, Henri G Franquelim, Sergei Korneev, Christian Schroeer, Petra Schwille, Dirk Trauner, Joost CM Holthuis and James A Frank is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.