Excitatory amino acid transporters (EAATs) are important in many physiological processes and crucial for the removal of excitatory amino acids from the synaptic cleft.*
In the article “Millisecond dynamics of an unlabeled amino acid transporter “ Tina R. Matin, George R. Heath, Gerard H. M. Huysmans, Olga Boudker and Simon Scheuring develop and apply high-speed atomic force microscopy line-scanning (HS-AFM-LS) combined with automated state assignment and transition analysis for the determination of transport dynamics of unlabeled membrane-reconstituted GltPh, a prokaryotic EAAT homologue, with millisecond temporal resolution.*
Among the bulk and single-molecule techniques, high-speed atomic force microscopy ( HS-AFM ) stands out with its ability to provide real-time structural and dynamical information of single molecules. HS-AFM images label-free molecules under close-to-physiological conditions with ~0.1 nm vertical and ~1 nm lateral imaging resolution. Furthermore, HS-AFM has typically ~100 ms temporal resolution, giving access to structure–dynamics relationship of proteins, though the achievable imaging speed depends on sample characteristics like scan size and surface corrugation.
Recently in a quest to achieve higher temporal resolutions, the authors of the cited article used HS-AFM line scanning (HS-AFM-LS) for the analysis of single-protein dynamics. *
Line scanning, using a conventional AFM, has been used to study protein–protein interactions earlier. In HS-AFM-LS, the slow-scan axis (y-direction) is disabled. Therefore, instead of imaging an x/y-area, the scientists scan over one horizontal x-line several hundreds to thousands of times per second, thus reaching millisecond temporal resolution. The topographical readouts of this line are stacked one after another, resulting in kymographs of the dynamical behavior of the molecules. Therefore, HS-AFM-LS has between 2 and 3 orders of magnitude higher temporal resolution than HS-AFM imaging and should allow the detection of fast transporter dynamics and possible intermediate states that have so far escaped kinetic characterization. *
All AFM images presented in this study were taken using a HS-AFM operated in amplitude modulation mode (with typical free and setpoint amplitudes, Afree = 1.0 nm and Aset = 0.9 nm, respectively using optimized scan and feedback parameters. NanoWorld Ultra-Short Cantilevers ( NanoWorld’s AFM probe series especially dedicated for High Speed Scanning) of the USC-F1.2-k0.15 type were used. In the presented experiments, four different buffer conditions were used. *
As the authors state in their article they find that GltPh transporters can operate much faster than previously reported, with state dwell-times in the 50 ms range, and report the kinetics of an intermediate transport state with height between the outward- and inward-facing states. Transport domains stochastically probe transmembrane motion, and reversible unsuccessful excursions to the intermediate state occur. The presented approach and analysis methodology are generally applicable to study transporter kinetics at system-relevant temporal resolution.*
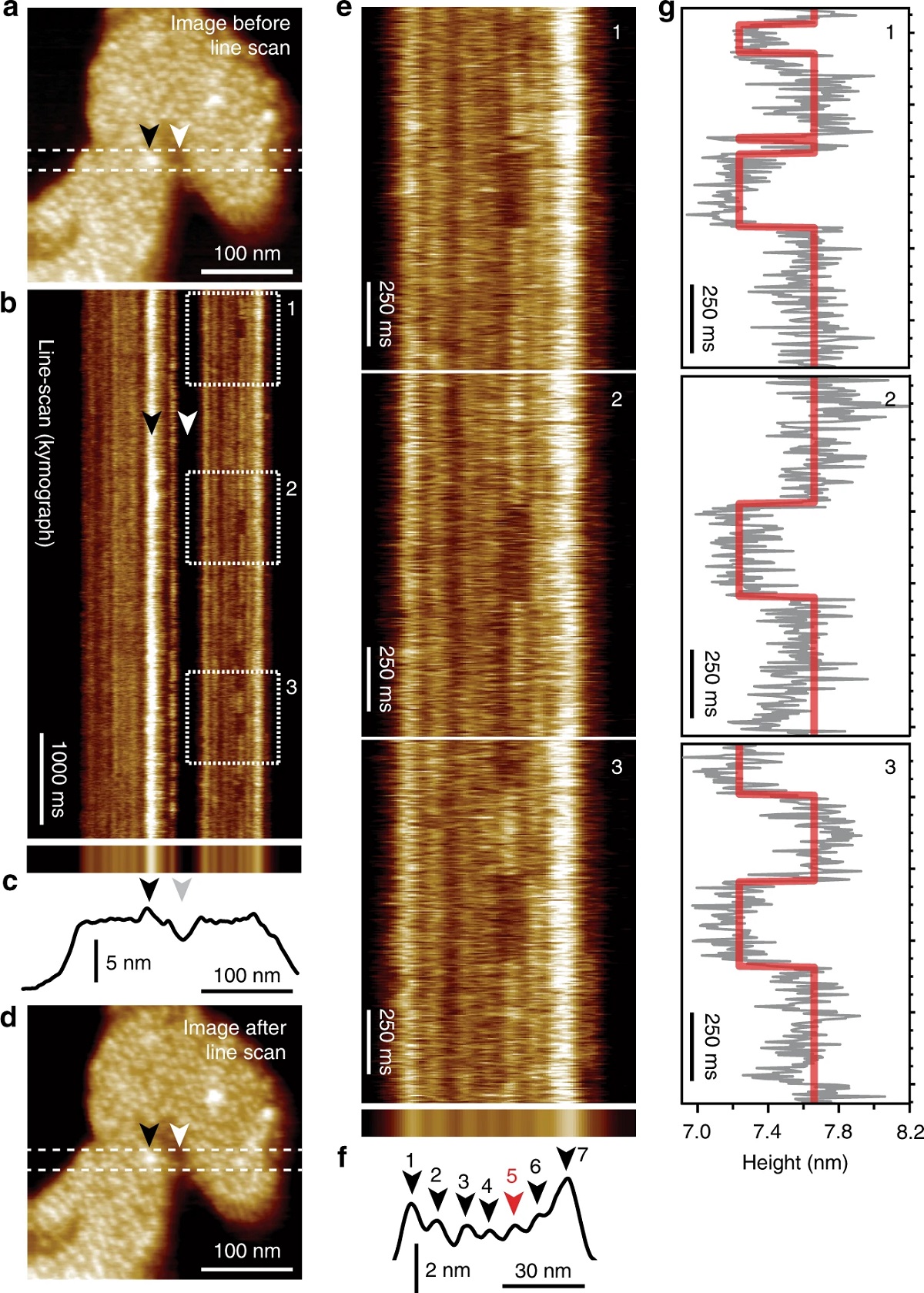
HS-AFM line scanning (HS-AFM-LS): millisecond temporal resolution of unlabeled transporter dynamics.:
a HS-AFM image of a membrane packed with GltPh exposing the extracellular face before HS-AFM-LS (apo condition: 20 mM Tris-HCl, pH7.5, 150 mM KCl). Dashed lines indicate the position of the central scan line where subsequent HS-AFM-LS is performed. b Six seconds of a HS-AFM-LS kymograph with 3.3 ms line acquisition speed. Each transporter domain appears as a vertical line. c Projection (top) and height profile (bottom) of b. d HS-AFM image after HS-AFM-LS. The lateral position of recognizable features in a–d are indicated by arrowheads. e One second high-magnification views of dashed regions 1, 2, and 3 in b. Transport domain excursions to the inward-facing state appear as dark dwells along the vertical time axis. f Projection (top) and height profile (bottom) of e. Arrowheads indicate the position of the seven protomers in the kymograph (red: active protomer #5). g Height/time traces (gray) and state fits (red) of the active domain (protomer #5) in e. This figure is representative of the experimental sequence for the >50 replicates analyzed in this work.
*Tina R. Matin, George R. Heath, Gerard H. M. Huysmans, Olga Boudker and Simon Scheuring
Millisecond dynamics of an unlabeled amino acid transporter
Nature Communications volume 11, Article number: 5016 (2020)
DOI: https://doi.org/10.1038/s41467-020-18811-z
Please follow this external link to read the full article: https://rdcu.be/cbuOU
Open Access : The article “Millisecond dynamics of an unlabeled amino acid transporter” by Tina R. Matin, George R. Heath, Gerard H. M. Huysmans, Olga Boudker and Simon Scheuring is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/.



