Although DNA origami nanostructures have found their way into numerous fields of fundamental and applied research, they often suffer from rather limited stability when subjected to environments that differ from the employed assembly conditions, that is, suspended in Mg2+-containing buffer at moderate temperatures.*
In the article “Cryopreservation of DNA Origami Nanostructures” Yang Xin, Charlotte Kielar, Siqi Zhu, Christoph Sikeler, Xiaodan Xu, Christin Möser, Guido Grundmeier, Tim Liedl, Amelie Heuer-Jungemann, David M. Smith and Adrian Keller investigate means for efficient cryopreservation of 2D and 3D DNA origami nanostructures and, in particular, the effect of repeated freezing and thawing. It is found that, while the 2D DNA origami nanostructures maintain their structural integrity over at least 32 freeze–thaw cycles, ice crystal formation makes the DNA origami gradually more sensitive toward harsh sample treatment conditions. *
The cryoprotectants glycerol and trehalose are found to efficiently protect the DNA origami nanostructures against freeze damage at concentrations between 0.2 × 10−3and 200 × 10−3m and without any negative effects on DNA origami shape. This work thus provides a basis for the long-term storage of DNA origami nanostructures, which is an important prerequisite for various technological and medical applications. *
NanoWorld Ultra-Short Cantilevers for High-Speed AFM USC-F0.3-k0.3 were used for the AFM imaging in liquid of the DNA origami sample described in this article.
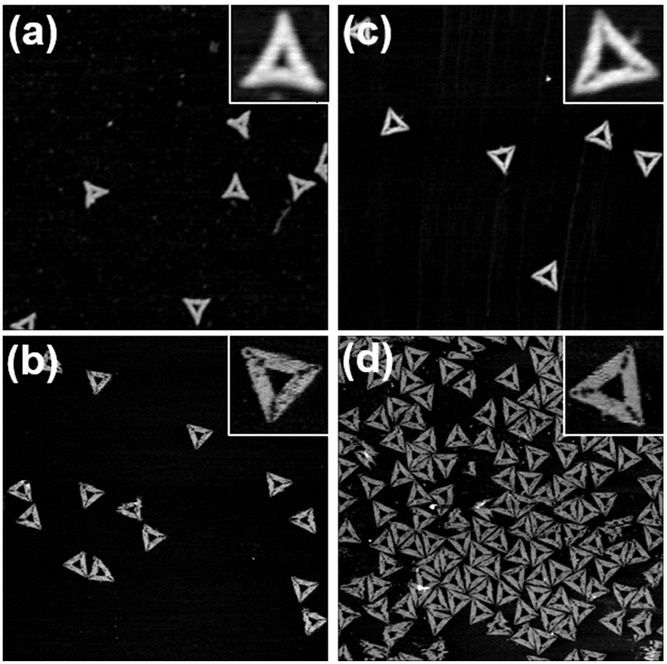
AFM images of triangular DNA origami nanostructures after 32 freeze–thaw cycles measured a) in air and b) in liquid. AFM images of triangular DNA origami nanostructures assembled from scaffold and staple strands that were subjected to 32 freeze–thaw cycles measured c) in air and d) in liquid. Images have a size of 1.5 × 1.5 μm2 and height scales are 2.3 nm.
*Yang Xin, Charlotte Kielar, Siqi Zhu, Christoph Sikeler, Xiaodan Xu, Christin Möser, Guido Grundmeier, Tim Liedl, Amelie Heuer-Jungemann, David M. Smith and Adrian Keller
Cryopreservation of DNA Origami Nanostructures
Small 2020, 16, 1905959
DOI: 10.1002/smll.20190595
Please follow this external link to read the full article: https://onlinelibrary.wiley.com/doi/pdf/10.1002/smll.201905959
Open Access The article “ Cryopreservation of DNA Origami Nanostructures “ by Yang Xin, Charlotte Kielar, Siqi Zhu, Christoph Sikeler, Xiaodan Xu, Christin Möser, Guido Grundmeier, Tim Liedl, Amelie Heuer-Jungemann, David M. Smith and Adrian Keller is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.


