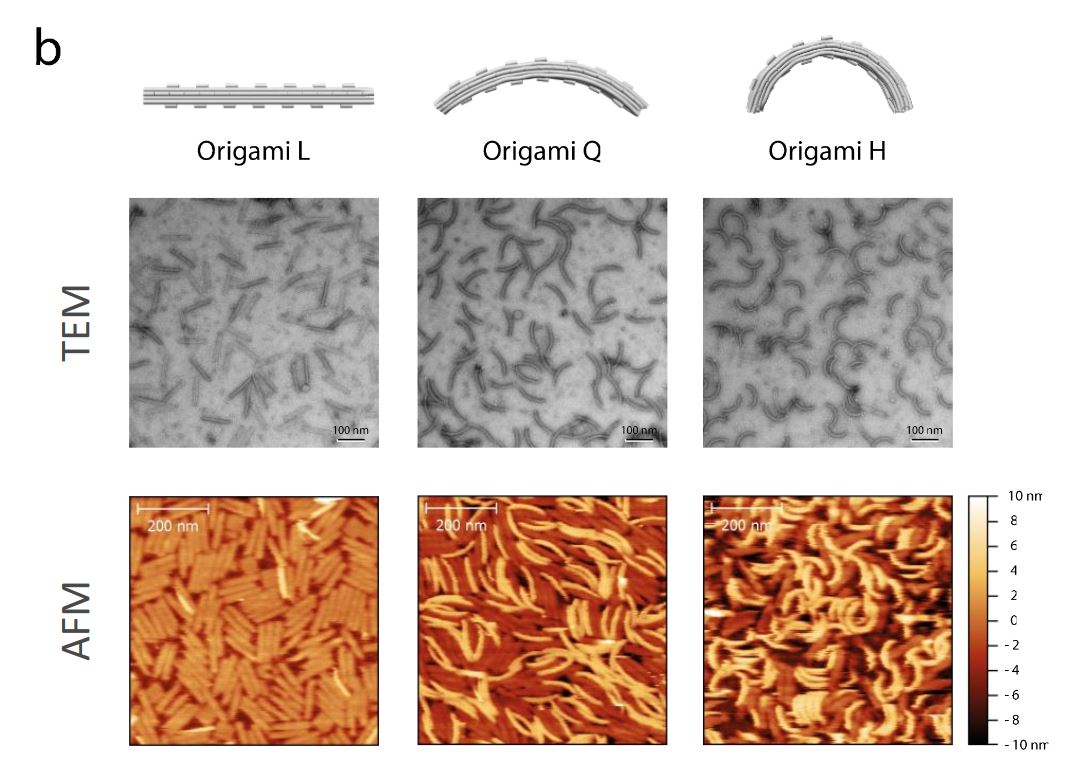
Retroviral integration, the process of covalently inserting viral DNA into the host genome, is a point of no return in the replication cycle. Yet, strand transfer is intrinsically iso-energetic and it is not clear how efficient integration can be achieved.*
In the article “The free energy landscape of retroviral integration” published in Nature Communications Willem Vanderlinden, Tine Brouns, Philipp U. Walker, Pauline J. Kolbeck, Lukas F. Milles, Wolfgang Ott, Philipp C. Nickels, Zeger Debyser and Jan Lipfert use biochemical assays, atomic force microscopy (AFM), and multiplexed single-molecule magnetic tweezers (MT) to study tetrameric prototype foamy virus (PFV) strand-transfer dynamics.*
Their finding that PFV intasomes employ auxiliary-binding sites for modulating the barriers to integration raises the question how the topology of higher-order intasomes governs integration of pathogenic retroviruses, most notably HIV. The single-molecule assays developed in this work are expected to be particularly useful to further unravel the complexity of this important class of molecular machines.*
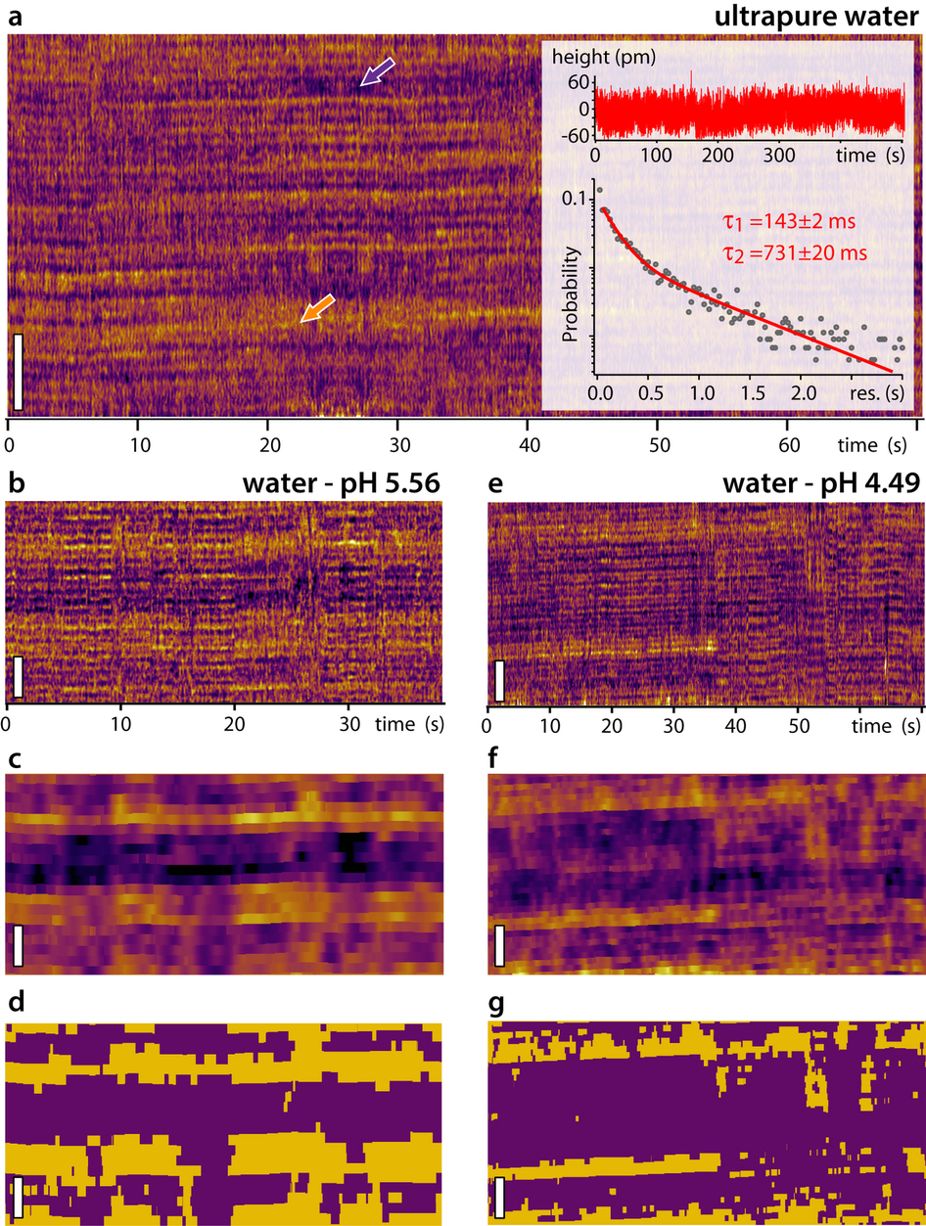
The AFM images were recorded in amplitude modulation mode under ambient conditions and by using NanoWorld high resolution SuperSharpSilicon™ SSS-NCH cantilevers ( resonance frequency ≈300 kHz; typical end-radius 2 nm; half-cone angle <10 deg). Typical scans were recorded at 1–3 Hz line frequency, with optimized feedback parameters and at 512 × 512 pixels.*

(please refer to the full article for the complete figure 2 https://rdcu.be/b0R63 ) :
e AFM image of intasomes incubated briefly (2 min) with supercoiled plasmid DNA, depicting a branched complex as found in ~50% of early complexes.
f AFM image of a bridging complex that dominates (~80%) the population of complexes at longer (>45 min) incubation.
g AFM image of a gel-purified STC
*Willem Vanderlinden, Tine Brouns, Philipp U. Walker, Pauline J. Kolbeck, Lukas F. Milles, Wolfgang Ott, Philipp C. Nickels, Zeger Debyser, Jan Lipfert
The free energy landscape of retroviral integration
Nature Communications volume 10, Article number: 4738 (2019)
DOI: https://doi.org/10.1038/s41467-019-12649-w
Please follow this external link to read the full article: https://rdcu.be/b0R63
Open Access The article “The free energy landscape of retroviral integration“ by Willem Vanderlinden, Tine Brouns, Philipp U. Walker, Pauline J. Kolbeck, Lukas F. Milles, Wolfgang Ott, Philipp C. Nickels, Zeger Debyser and Jan Lipfert is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.