Mucosal immunoglobulins comprise mainly secretory IgA antibodies (SIgAs), which are the major contributor to pathogen-specific immune responses in mucosal tissues. SIgAs exist as mainly dimers and tetramers and play critical roles in mucosal immune responses against influenza.*
Detailed characterization of these anti-viral SIgA is important for better understanding of the mechanisms underlying anti-viral immunity.*
In their article “IgA tetramerization improves target breadth but not peak potency of functionality of anti-influenza virus broadly neutralizing antibody” Saito S, Sano K, Suzuki T, Ainai A, Taga Y, Ueno T, et al. (2019) describe a means of generating a recombinant tetrameric monoclonal SIgA to enable exhaustive characterization of tetrameric SIgAs.
The tetrameric monoclonal SIgA possessing variable regions of anti-influenza viruses broadly neutralizing antibody show that tetramerization of SIgA improves target breadth, but not the peak potency, of their anti-viral functions.*
These results broaden the knowledge about the fundamental role of SIgA tetramerization in anti-viral humoral response at the human respiratory mucosa.*
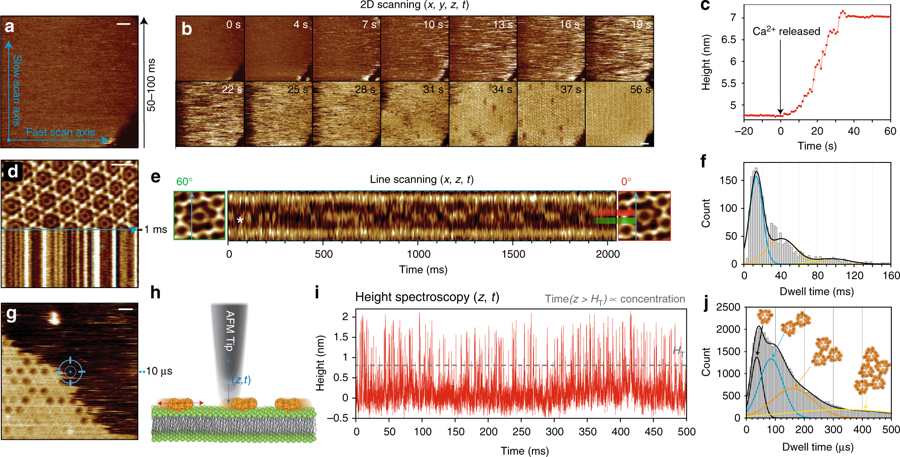
The high speed atomic force microscopy ( HS-AFM ) experiments mentioned in the article were performed using a NanoWorld Ultra-Short Cantilever USC-F1.2-k0.15.

Fig 1. Production of recombinant tetrameric monoclonal SIgAs from ”
IgA tetramerization improves target breadth but not peak potency of functionality of anti-influenza virus broadly neutralizing antibody ” by
Saito S, Sano K, Suzuki T, Ainai A, Taga Y, Ueno T, et al. (2019) :
(A) Recombinant monoclonal IgA antibodies purified from the culture supernatant of cells co-transfected with A1+L (left upper), A1+L+J (left lower), A1+L+J+SC (right upper), or A2m2+L+J+SC (right lower), were subjected to size exclusion chromatography (SEC) analysis. A chromatogram showing absorbance at 280 nm revealed three major peaks: peak A (retention volume around 10.4 ml), peak B (retention volume around 9.3 ml), and peak C (retention volume around 8.4 ml). Data are representative of three independent experiments. (B) SDS-PAGE and BN-PAGE analysis of IgG and IgA1/IgA2m2 in each peak fraction (peak A, B, and C) purified from cells co-expressing SC (A1+L+J+SC or A2m2+L+J+SC). (C, D, E) High-mass MALDI-TOF MS analysis of the each peak fraction containing recombinant IgA1 purified from the culture supernatant of cells transfected with A1, L, J, and SC. (C) One main peak (arrow) corresponding to monomer (Mo) was detected in the peak A fraction. (D) Two main peaks (arrows) corresponding to a dimer (Di) and a di-cation dimer (Di2+) were detected in the peak B fraction. (E) Three main peaks (arrows) corresponding to a tetramer (Te), trimer (Tr), and di-cation tetramer (Te2+) were detected in the peak C fraction. (F, G) High-mass MALDI-TOF MS analysis of the each peak fraction of recombinant IgA2m2 purified from the culture supernatant from cells transfected with A2m2, L, J, and SC. (F) One main peak (arrow) corresponding to a monomer (Mo) was detected in the peak A fraction. (G) Three main peaks (arrows) corresponding to a tetramer (Te), a trimer (Tr), and a di-cation tetramer (Te2+) were detected in the peak C fraction. (H) Quantification of the amount of each subunit within the peak B or C fraction of recombinant SIgA1 or SIgA2m2 antibodies purified from the culture supernatant of cells transfected with A1/L/J,/SC or A2m2/L/J/SC using LC-MS with stable isotope-labeled standard peptides. The abundance of each subunit to that of J chain is expressed as a ratio. Data are expressed as box-and-whisker plot with minimum, maximum, median, upper and lower quartiles (n = 6–7). (I) HS-AFM image of peak C derived from a recombinant SIgA1 (A1Te) or SIgA2m2 (A2m2Te) antibody purified from the culture supernatant of cells transfected with A1/L/J/SC or A2m2/L/J/SC. Scale bar, 20 nm.
*Shinji Saito , Kaori Sano , Tadaki Suzuki , Akira Ainai, Yuki Taga, Tomonori Ueno, Koshiro Tabata, Kumpei Saito, Yuji Wada, Yuki Ohara, Haruko Takeyama, Takato Odagiri, Tsutomu Kageyama, Kiyoko Ogawa-Goto, Pretty Multihartina, Vivi Setiawaty, Krisna Nur Andriana Pangesti, Hideki Hasegawa
IgA tetramerization improves target breadth but not peak potency of functionality of anti-influenza virus broadly neutralizing antibody
PLoS Pathog 15(1): e1007427.
DOI: https://doi.org/10.1371/journal.ppat.1007427
Please follow this external link for the full article: https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1007427
Open Access: The article « IgA tetramerization improves target breadth but not peak potency of functionality of anti-influenza virus broadly neutralizing antibody » by Saito S, Sano K, Suzuki T, Ainai A, Taga Y, Ueno T, et al. (2019) is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.