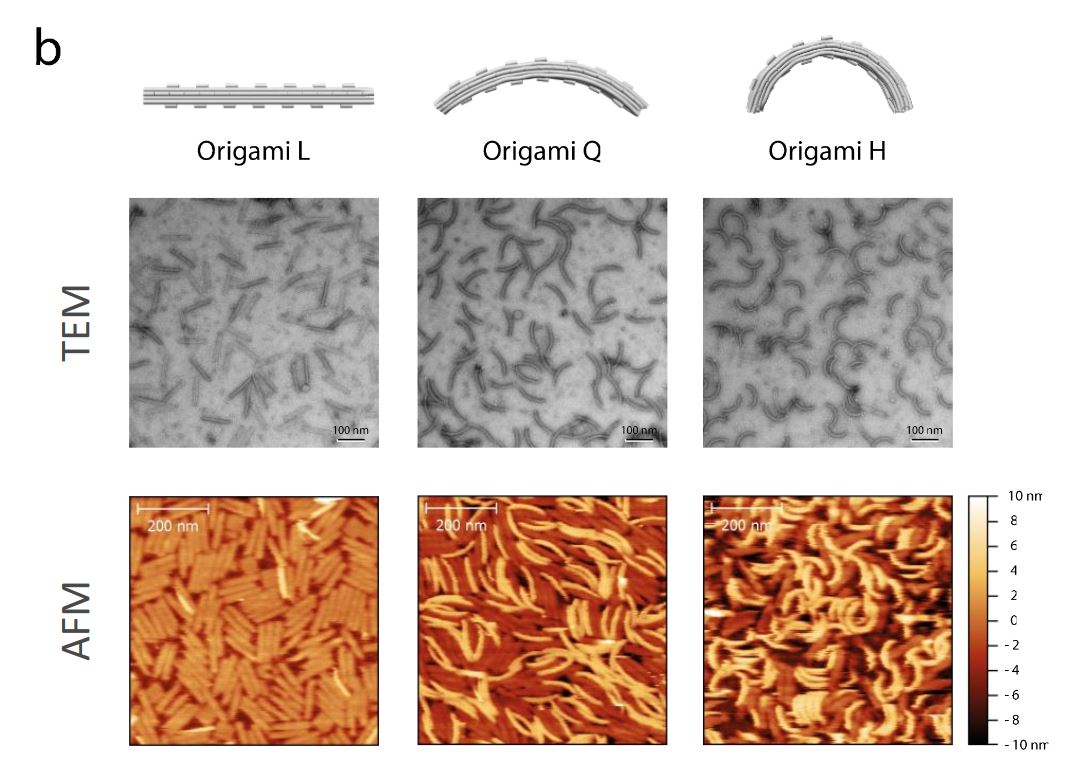
In the article “Direct observation of dynamic interaction between a functional group in a single SBR chain and an inorganic matter surface” Ken-ichi Shinohara and Yuu Makida use atomic force microscopy (AFM) video imaging to closely investigate the behaviour of functionalized and unmodified styrene-butadiene rubber (SBR), as models for tire rubber, on mica surfaces.
“Using AFM video imaging, we tracked the behavior of individual SBR polymer chains on mica surfaces to reveal how polymer modification affects the interaction of SBR with mica surfaces. We measured the diffusion coefficients and spring constants of single SBR polymer chains for the first time, demonstrating that it is possible to parameterize the relationship between the molecular dynamic structure of a polymer and rubber properties of the vulcanized compound.”*
NanoWorld Ultra-Short Cantilevers (USC) for Fast-/High-Speed AFM ( USC-F1.2-k0.15 ) were used

*Ken-ichi Shinohara & Yuu Makida
Direct observation of dynamic interaction between a functional group in a single SBR chain and an inorganic matter surface
Nature Scientific Reports, volume 8, Article number: 13982 (2018)
DOI: https://doi.org/10.1038/s41598-018-32382-6
For the full article please follow this external link: https://rdcu.be/bbERH
The article “Direct observation of dynamic interaction between a functional group in a single SBR chain and an inorganic matter surface” by Ken-ichi Shinohara & Yuu Makida is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.